pKALM: Accurate and Rapid Protein pKa Prediction: Protein Language Models Reveal the Sequence-pKa Relationship
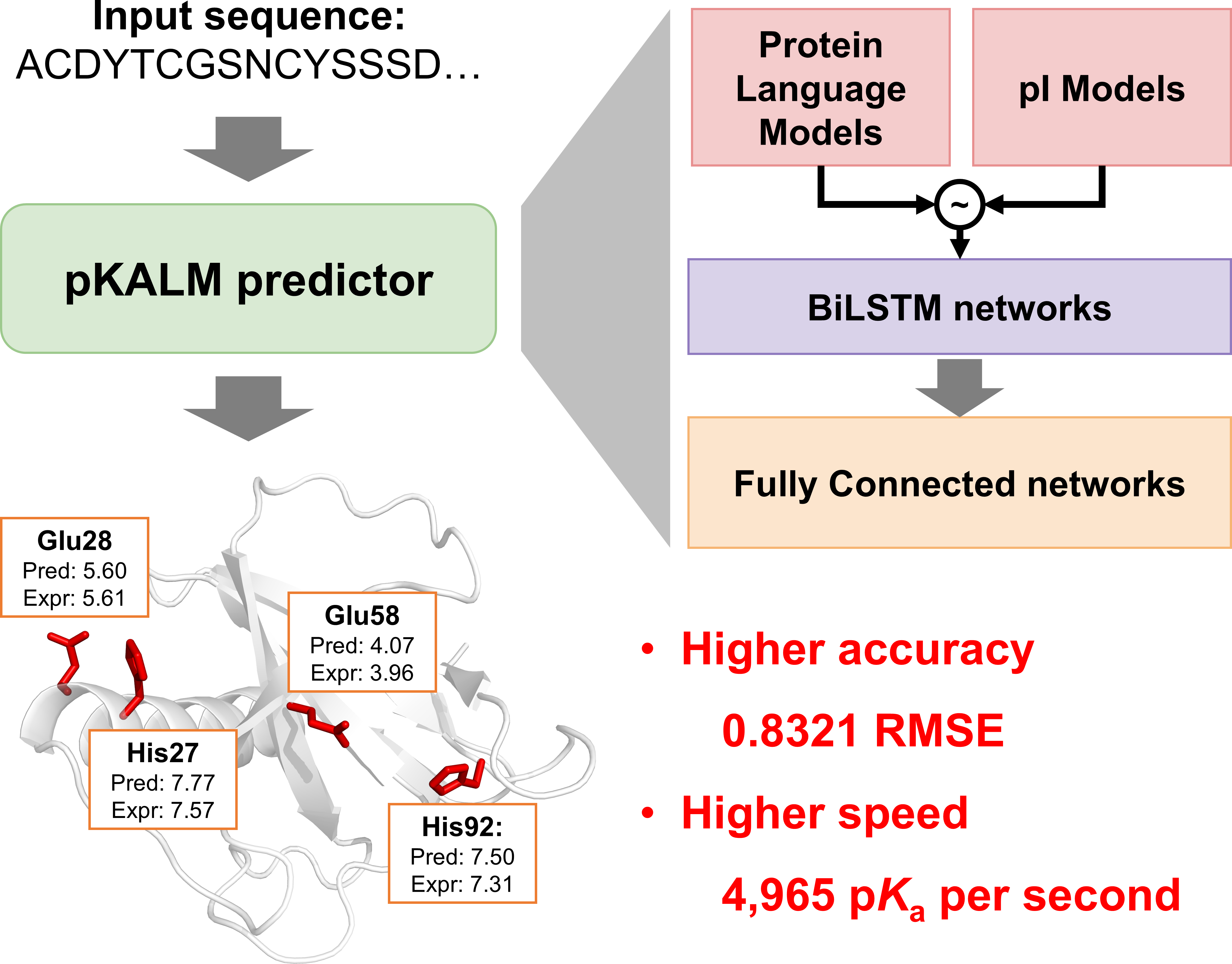
pKALM is a web server for predicting protein pKa values from amino acid sequences, based on protein language model and transfer learning. It achives high accuracy and speed compared to existing methods. It supports the pKa prediction of eight ionizable groups: N-terminal, C-terminal, and six ionizable side chains of Asp, Glu, His, Lys, Cys, and Tyr, with 0.8321 RMSE and a speed of 4,961 pKa / sec. [ GitHub | Paper | License ]
Input sequence is in
FASTA format. It should be less than 2,000 amino acids. For the prediction of longer sequences, please contact us.
There is a variant of pKALM, called pKALMs, which can predict largely shifted pKa values. pKALMs was trained on the simulated data rather than the experimental data. Users can choose either pKALM or pKALMs for the prediction.
Prediction Results
The prediction is quite fast and the results is a CSV-formatted table with 5 columns:
- The first column is the sequence ID.
- The second column is the index of the ionizable group.
- The third column is the name of the ionizable group.
- The fourth column is the predicted pKa shift values.
- The fifth column is the predicted pKa values.
Significantly shifted pKa values are highlighted in yellow. The predicted pI are also attached at the end of the table.
References
Users are kindly requested to utilize the following citation when referencing this method:
- Shijie Xu and Akira Onoda, Accurate and Rapid Prediction of Protein pKa: Protein Language Models Reveal the Sequence-pKa Relationship." bioRxiv (2024): 2024-09. DOI: 10.1101/2024.09.16.613101
Please contact
shijie.xu@ees.hokudai.ac.jp for any questions.
Changelogs
- 2024/10/22: First release.