PsiPartition: Improved Site Partitioning for Genomic Data by Parameterized Sorting Indices and Bayesian Optimization
PsiPartition is a phylogenetic tool using Bayesian optimization to obtain more accurate reconstructions, especially for large genomic data with more site heterogeneity. It is designed with a parameterized sorting index which is further optimized by Bayesian optimization, to partition sites into different categories. Partitioned models in main-stream phylogenetic softwares (e.g., IQ-TREE) can be used to analyze the categories. [ GitHub | Paper ]
Introduction
DNA (deoxyribonucleic acid) is the blueprint of life. It carries the instructions to make proteins, which are essential for all living things. These instructions are written using a code made of three-letter "words" called codons (Figure 1). The same codon code is used by almost all life forms, showing how all living things are connected. However, the code is not one-to-one, meaning that multiple codons can codee for the same amino acid. This redundancy is called "degeneracy" and is thought to be a result of evolution. The difference of code for the same amino acids usually happens at the third position of the codon, which is called the "synonymous" site. In contrast, the first and second positions are called "non-synonymous" sites. Such as difference means that the third position is less important for the protein structure and function, and is under less selective pressure.
 Figure 1: The genetic code. The code is made of three-letter "words" called codons. Each codon codes for an amino acid, which are the building blocks of proteins. CC https://ib.bioninja.com.au/genetic-code/.
Figure 1: The genetic code. The code is made of three-letter "words" called codons. Each codon codes for an amino acid, which are the building blocks of proteins. CC https://ib.bioninja.com.au/genetic-code/.
The difference in selective pressure between synonymous and non-synonymous sites are considered in the partitioned models in phylogenetic inference. These models assume that different sites in the sequence alignment have different evolutionary rates, and use different substitution matrices for different sites. The partitioned models can improve the accuracy of phylogenetic inference, especially for large genomic data with more site heterogeneity. However, the partitioned models require the user to specify the number of partitions and the sites in each partition. This is a challenging task, as the user needs to have prior knowledge of the data to make the partitioning.
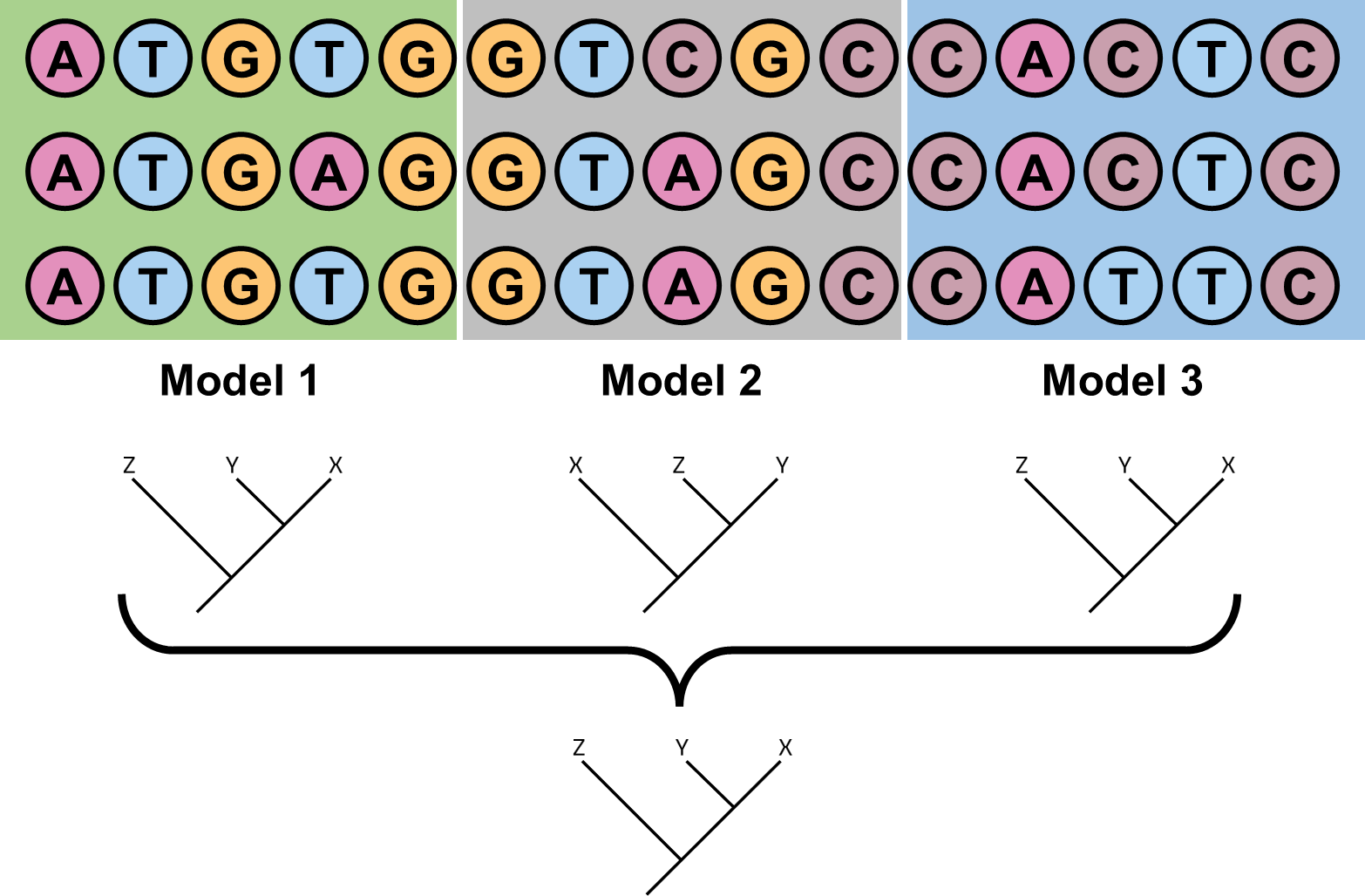 Figure 2: Partitioned models in phylogenetic inference. The models assume that different sites in the sequence alignment have different evolutionary rates, and use different substitution matrices for different sites.
Figure 2: Partitioned models in phylogenetic inference. The models assume that different sites in the sequence alignment have different evolutionary rates, and use different substitution matrices for different sites.
How to use
Step 1. Preparation
Before using PsiPartition, please prepare the following things:
- A sequence alignment in FASTA format. Suppose you have a query sequence and you want to infer its phylogenetic relationship with other sequences. You need to search for homologous sequences from the database such as UniProt and align them by using software such as COBALT.
- Phylogenetic software. PsiPartition is based on partitioned models, and it self does not perform phylogenetic inference. We use IQ-TREE as our host software. Click the link and unzip the files into some folder. You can type './bin/iqtree2.exe -s example.phy' to test if it works.
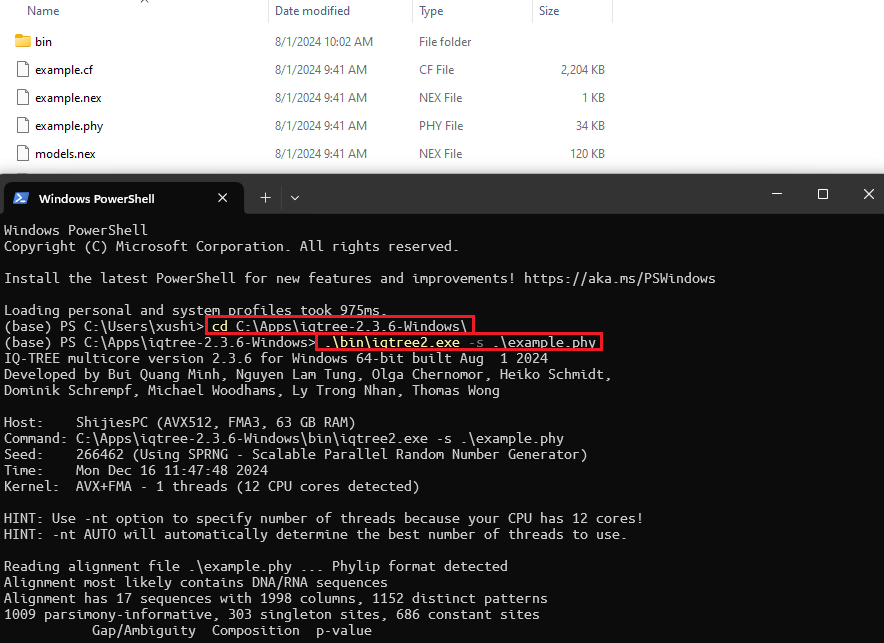 Figure 1: IQ-TREE phylogenetic software. The software is used to infer phylogenetic trees from sequence alignments.
Figure 1: IQ-TREE phylogenetic software. The software is used to infer phylogenetic trees from sequence alignments.
- Python. PsiPartition is written in Python, so you need to have Python installed on your computer. You can download Python from here.
- PsiPartition. Download the PsiPartition software from here. Also unzip it somewhere. Go to the folder and type `pip install -r requirements.txt` to install the required packages.
- A wandb account. PsiPartition uses Weights & Biases to log the optimization process. You need to sign up for an account and get your API key.
Step 2. Run PsiPartition
After you have prepared the above things, you can run PsiPartition by following the steps below:
python PsiPartition_wandb.py --msa MSA File --format fasta or phylip --alphabet dna or aa --max_partitions max_partitions --n_iter number of iterations
where the arguments are:
- --msa: The path to the sequence alignment file in FASTA format.
- --format: The format of the alignment file. It can be either 'fasta' or 'phylip'.
- --alphabet: The alphabet of the sequences. It can be either 'dna' or 'aa'.
- --max_partitions: The maximum number of partitions to be optimized.
- --n_iter: The number of iterations for Bayesian optimization.
This will take some time to run, depending on the size of the alignment and the iterations. After the optimization is done, PsiPartition will output the optimized partitioning scheme. You can then use this partitioning scheme to analyze the data with partitioned models in IQ-TREE. An example output is shown below:
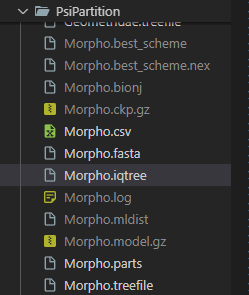 Figure 2: Example output of PsiPartition.
The `*.iqtree` file contains the reconstructed phylogenetic tree. You can visualize the tree using software such as iTOL. In addition, the file `*.parts` contains the optimized partitioning scheme. You can use this file to analyze the data with partitioned models in IQ-TREE. For example, you can run the following command to infer a phylogenetic tree with the optimized partitioning scheme:
Figure 2: Example output of PsiPartition.
The `*.iqtree` file contains the reconstructed phylogenetic tree. You can visualize the tree using software such as iTOL. In addition, the file `*.parts` contains the optimized partitioning scheme. You can use this file to analyze the data with partitioned models in IQ-TREE. For example, you can run the following command to infer a phylogenetic tree with the optimized partitioning scheme:
./bin/iqtree2.exe -s example.phy -spp example.parts
References
Users are kindly requested to utilize the following citation when
referencing this method:
-
Xu, Shijie, and Akira Onoda. "PsiPartition: Improved Site Partitioning for
Genomic Data by Parameterized Sorting Indices and Bayesian Optimization."
Journal of Moelecular Evolution.
Please contact
shijie.xu@ees.hokudai.ac.jp
for any questions.
Changelogs
- 2024/08/08: First release.
- 2024/12/16: Added the guide on how to use PsiPartition.